- Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets
- 관리자 |
- 2025-07-08 17:25:39|
- 7
- 2025-07-08 17:25:39|
ㅇ [Title] Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets
ㅇ [Journal] Proceedings of the National Academy of Sciences of the United States of America (PNAS)
ㅇ [Author] JinA Lim#, Hae Deok Jung#, Soo Young Park, Moonhyeon Jeon, Da Sol Kim, Ryeongeun Cho, Dohyun Han, Han Suk Ryu, Yoosik Kim*, Hyun Uk Kim*
#: co-first author, : corresponding author
ㅇ [Abstrct]
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such
approaches can often lead to even stronger resistance. Here, we develop a computational framework that predicts metabolic gene targets capable of reverting the metabolic state of
drug-resistant cells to that of drug-sensitive parental cells, thereby sensitizing the resistant cells. The computational framework performs single-gene knockout simulation of genome-scale
metabolic models that predicts genome-wide metabolic flux distribution in drug-resistant cells, and clusters the resulting knockout flux data using uniform manifold approximation and
projection, followed by k-means clustering. This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
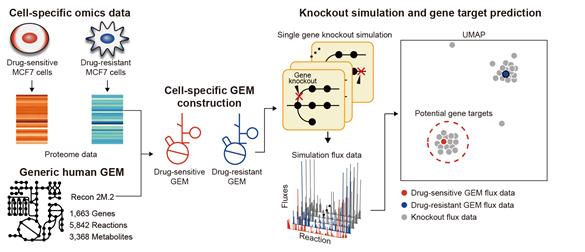
Framework of reconstructing and simulating cell-specific GEMs to predict gene targets that can sensitize the drug-resistance breast cancer cells.
| 첨부파일 |
|
|---|

